Bio-Chemistry-Assignment-NMR-COSY-NOESY-HKL-Calculations
Author: Ellie Cross
At: July 21, 2023
Question#1
Components of Nuclear Magnetic Resonance
Nuclear magnetic resonance (NMR) is a marvel wherein cores in a solid, consistent appealing field are irritated by a delicate influencing alluring field and respond by making an electromagnetic sign with a repeat typical for the appealing field at the centre.
The Magnet: The capacity of a NMR instrument depends upon the extent and homogeneity of the static attractive field and on the drag size of the magnet.
The Gradient System: The age of attractive reverberation pictures depends on properly utilising beat attractive field inclinations. These angles are created similarly to those delivered by the shim curls.
The Transmitter: The transmitter creates radiofrequency beats of the fitting recurrence, force, shape, and timing. It contains a recurrence generator, and a waveform generator to shape the beats.
The Radiofrequency coil: Radiofrequency loops are utilized to communicate the B1 field into the area of interest and identify the subsequent sign.
The Receiver: A cutting-edge computerized recipient’s plan is based on a simple to the advanced converter, which tests the simple NMR sign and converts it into the advanced arrangement.
The Computer: The PC has a wide scope of capabilities. Its primary capacities are to control the radiofrequency and field angle beats, aggregate the information, and measure and show the information.
COSY and NOESY Spectra
The primary phase of the successive tasks includes the recognizable proof of frameworks of turn-coupled resonances compared to singular amino corrosive deposits. The irregular loop substance move values for resonances in the regular amino acids are appeared. Large numbers of the amino acids have exceptional turn framework geographies and will offer ascent to one-of-a-kind examples of cross tops in a Cosy range. Nonetheless, in many proteins, there is some level of cover that will probably increment with the protein’s size. When cover happens, unambiguous turn framework tasks are certainly not generally conceivable based on the Cosy range alone, and other 2D datasets are required. It will be demonstrated later that the investigation of NOE information for the second phase of successive tasks depends most vigorously on the HN and Hα resonances.
NOESY Spectra:
The second stage of assignment involves the particular buildup in the protein grouping. This is accomplished by connecting an amino corrosive turn framework with the turn frameworks of its neighbouring deposits in the arrangement. It depends on the through-space networks seen in NOESY spectra. The most valuable NOE impacts for successive task were found to include the Hα of buildup I and the HN of buildup i+1, dαN(i,i+1), the HN’s of deposits I and i+1, dNN(i,i+1), and the Hβ of buildup I and the HN of buildup i+1, dβN(i,i+1). The powers of these NOE impacts rely upon the twist points ψ, φ and ψ, and χ1 and ψ, separately. This twist point reliance implies that explicit consecutive NOE impacts portray particular kinds of auxiliary structure. In the all-inclusive spine structure, normal for β-sheet, the distance dαN(i,i+1) is short, though dNN(i,i+1) is longer. The distance dNN(i,i+1) is short in helical structure, though dαN(i,i+1) is longer. NOE impacts between HN, Hα also, and Hβ resonances are not, be that as it may, limited to adjoining deposits of the grouping.
Question#2 Patterson Map
In 1935, Patterson demonstrated that the obscure stage data in the condition for electron thickness:
ρ(xyz) = 1/V ∑h ∑k ∑l |F(hkl)| exp[iα(hkl)] exp[-2πi(h x + k y + l z)]
can be eliminated on the off chance. We do the summation on F2(hkl) and agree that the totality of the phases is 0. This is the work of Patterson:
P(uvw) = 1/V ∑h ∑k ∑l |F(hkl)|2 exp[-2πi(h u + k v + l w)]
Patterson suggested that this work contrasts the condition of a “top” at (u,v,w) with the vectors between two molecules (at (x1,y1,z1) and (x2,y2,z2) in the gem grid with the end target of:
u = x1 – x2 v = y1-y2 w = z1-z2
Note that a “map” refers to a 3-dimensional appropriation of maxima or “tops” within a unit cell or a progression of unit cells concerning crystallography. For a cell of a unit containing N
the related Patterson guide will display N2 tops, which can confuse things essentially. There are N tops at the cause, speaking to the vector between each molecule and itself, and N(N1) tops inside the unit cell. This can bring about an extremely stopped guide and difficult to translate. Note that the Patterson map is centrosymmetric, further stopping up cells for non-centrosymmetric networks. Bigger structures, for example, proteins, are ordinarily less unbending than more modest particles, and hence even in a precious stone, there will be huge confusion between unit cells. This restricts the goal of the diffraction example to normally 2 or 3 A˚ . Since these particles may likewise contain many iotas, an alternate way to deal with staging should be received. Data about the obscure stages might be acquired by rolling out a known improvement to the substance of the unit cell and estimating the impact on the diffraction design. It is typically conceivable to find the weighty molecules alone by using direct strategies or Patterson techniques. Once the area of the substantial molecules is known, the dispersing from those molecules might be determined both in size and stage.
Question#3 (a) Argand’s diagram
An Argand outline is a plot of complex numbers as focuses. in the intricate plane utilizing the x-pivot as the genuine hub and y-hub as the fanciful hub.
The IUCr portrays the “structure factor” Fhkl as “a mathematical limit that depicts the abundancy and time of a wave diffracted from valuable stone framework planes portrayed by Miller records h, k, l.where the whole is overall j particles in the unit cell, xj , yj and zj are the fragmentary bearings of the jth molecule, fj

is the dissipating element of the jth iota, and αhkl is the period of the structure factor. Additionally, │Fhkl│ 2 is corresponding to the power estimated in the investigation.
From Equation 3.3, we note that:
(1) For a known structure, the amplitudes and periods of the structure factors can be determined from the Fourier change of the structure. For an obscure structure, the amplitudes can be derived from the diffraction information, and the stages are lost during the diffraction try.
(2) The structure factor associates primary data in genuine and corresponding space. Each molecule in a unit cell in genuine space will add to the power of each appearance in proportional space.
(3) Structure elements can be portrayed as vectors in an Argand chart as appeared. Adding vector commitments from every molecule in a unit cell will give a last Fhkl.
(4) If the periods of all the Fhkl are known, the 3D electron thickness along the c-hub after picture handling by CRISP . In the event that the period of the most grounded reflection is left unaltered while its sufficiency is changed to 33% of the first adequacy, the primary underlying highlights of this projection are held.

Structure factor represented in argand diagram
B. Now, you may need to momentarily audit the prologue to vector portrayal of dispersing factors. The accompanying vector chart (Harker introduction) shows the connection among local and subsidiary dissipating factors. The goal of a staging test is to infer the obscure stage a (p) of every protein reflection Fp.
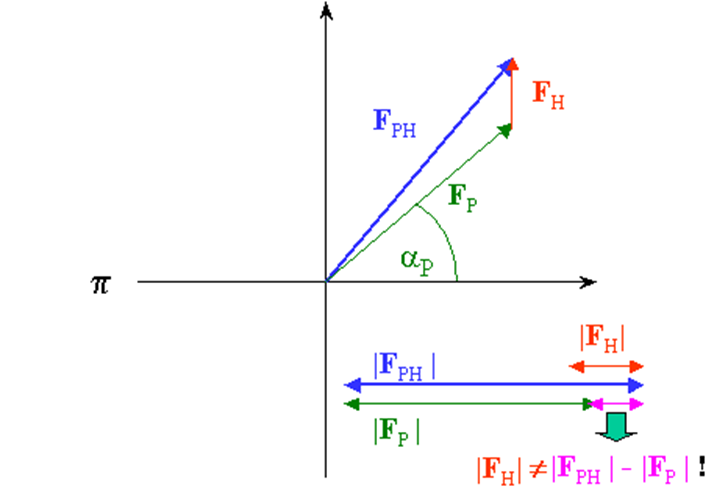
From our test, we know just the extent |Fph| (subsidiary) and |Fp| (protein), which can be spoken to in the intricate plane as a hover of range |Fph| and |Fp| , separately. On the off chance that we know both the extent and the period of Fh, we can draw the two circles balance by vector Fh and get 2 answers for conceivable stage esteems for Fp:
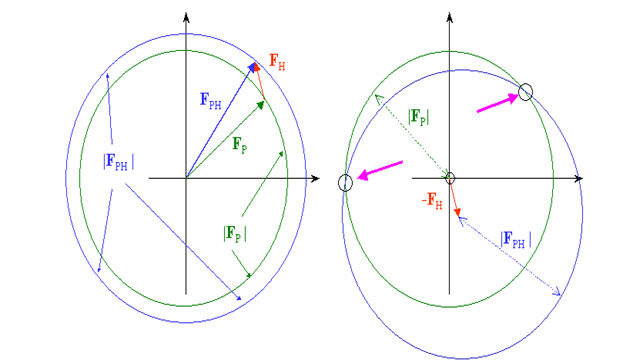
The stage and size of Fh can be determined effectively in the event that we know the places of a hefty metal. Now is certain that the best stage we can get from the 2 arrangements is the mean in the middle of the 2 prospects, and the stage blunder can be very enormous. In genuine cases, Fh is a lot more limited than my red vector and the stage blunder will be huge, fairly under 90 degrees.
To dispense with the stage uncertainty, we can set up a subsequent subordinate and rehash the method. We have now, from a certain point of view, a precise answer for the stage point of Fp. The hypothesis depends on 2 suspicions : a) ideal isomorphism and b) accurate hefty particle positions, neither of which are impeccably met, for pragmatic and test reasons in the primary case and for hypothetical reasons in the second. In our image, it implies that the staging circles may not meet precisely in one spot, and another subordinate might be important to improve the nature of the stages. The technique is accordingly called MIR, Multiple Isomorphous Replacement.

Question 4:
A: The quantity of a-helical and b-strand fragments was assessed from the portions of contorted helix ~ faD! In comparison, a warped strand ~ fbD! Determined by the study of CD spectra. These divisions compare with na deposits per a-helix ~aD! in guaranteed protein structure. Moreover, per b-strand, nb deposits ~bD!. Quantity of fragments a-helical ~Na! Using Na 5 (faD 3 Nres!0 na, and the quantity of b-strand parts ~Nb! was resolved using: Nb 5 ~ fbD 3 Nres!0nb, where Nres is the sum of protein deposits. The study show is portrayed by RMSDs ~d! In addition, relation coefficients ~r) between the X-beam and CD evaluations of auxiliary structure components for various optional structure functions, or gauges of Or then again, or then again, B-shaped strand ~Nb! Snippets. This is shown by dk and rk, where k is known to be one of the auxiliary underlying kinds. In general, the execution of the analysis for a given arrangement of optional structure divisions was governed by all in all thought regarding all auxiliary structure divisions, and d and r include these.
B: The hypothetical depiction of CD spectra of particles as extensive as DNA is extremely unpredictable, so the strategy can’t give primary data on the atoms at nuclear level. Therefore, CD spectroscopy is basically utilized observationally in investigations of DNA. Cd spectroscopy, be that as it may, has numerous preferences over different strategies for conformational examination. In the first place, it is very touchy, allowing work with DNA sums as low as 25 μg. The convergence of DNA can likewise be exceptionally low (20 μg/ml). This is worthwhile in investigations of tests of low dissolvability of those that will, in general, total under outrageous dissolvable conditions. Changes in the way length can be utilized to successfully modify DNA fixation by multiple significant degrees. Second, the contemplated atoms can be short yet, in addition, long, which is particularly significant with DNA. Third, the examples can, without much of a stretch, be titrated with different specialists that incite conformational isomerizations in DNA. This makes it conceivable to plan the entire conformational space of the examined particle, not simply a solitary structure.

a.Equilibrium dissociation constant for kdx
Kd= [X][Y]/[XY]
When X=Kd then Y=XY or equivalently Kd=[X][Y]/[Y+XY]
Kdx=3.2×107 x 8.0×105/8.0×105 + 3.2×107 x 8.0×105
Kdx= 1 M-1S-1
The equilibrium dissociation constant for kdy
Kd= [X][Y]/[XY]
When X=Kd then Y=XY or equivalently Kd=[X][Y]/[Y+XY]
Kdy=2.4×10-2x 1.6×10-4/1.6×10-4+ 3.84×10-6
Kdy= 0.0234 M-1S-1
b. The compound which has a higher value of equilibrium dissociation constant has a higher affinity to the target protein.
c. The compound X is likely to be a better therapeutic drug. There are various reasons why feeble restricting atoms have been ignored for quite a while as a hotspot for drug disclosure in natural sciences. To begin with, there is as yet a worldview in the psyches of numerous researchers expressing that impact and explicitness just come from drugs that quandary firmly to an objective atom. It has been imagined that a powerless fastener isn’t explicit to its objective and commonly shows high cross-reactivity to other restricting locales. Second, for powerless fondness particles, the measure of official to the objective can be seen as an issue, as it could be too low to even think about propelling a reaction. Be that as it may, if neighbourhood groupings of a powerless cover are sufficiently high, it can drive harmony, bringing about a significant bound ligand. Third, the vast majority of the current medication revelation models are unequipped for screening or dissecting feebly restricting medications or frail organic collaborations
References:
- Dictionary.iucr.org. 2021. Patterson Methods – Online Dictionary Of Crystallography
- Dictionary.iucr.org. 2021. Patterson Methods – Online Dictionary Of Crystallography.
- Rupp, B., 2021. Structure Factor Calculation.
- Roncador, G. et al. The European antibody network’s practical guide to finding and validating suitable antibodies for research. mAbs 8, 27–36, doi: 10.1080/19420862.2015.1100787 (2016).
- Redfield, C., 2021. Tight Vs. Loose Coupling Organizational Structure.
- Williams RW. Estimation of protein secondary structure from the laser Raman amide I spectrum. J Mol Biol. 1983 Jun 5;166(4):581–603.
- UKEssays. November 2018. NMR Spectrometer: Applications, Components and Functions.


